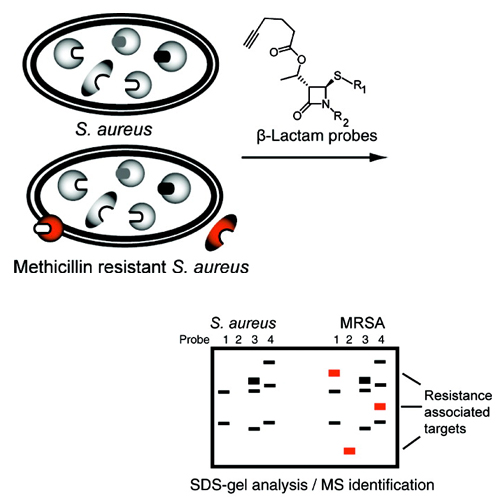
beta-Lactam Probes As Selective Chemical-Proteomic Tools for the Identification and Functional Characterization of Resistance Associated Enzymes in MRSA
08-Apr-2009
Journal of the American Chemical Society, 2009, DOI: 10.1021/ja901304n, published on 08.04.2009
JACS, online article
With the development of antibiotic resistant bacterial strains, infectious diseases have become again a life threatening problem. One of the reasons for this dilemma is the limited number and breadth of current therapeutic targets for which several resistance strategies have evolved over time. To identify resistance associated targets and to understand their function, activity, and regulation, we utilized a novel strategy based on small synthetic β-lactam molecules that were applied in activity based protein profiling experiments (ABPP) to comparatively profile in situ enzyme activities in antibiotic sensitive and resistant S. aureus strains (MRSA). Several enzyme activities which are unique to the MRSA strain including known resistant associated targets, involved in cell wall biosynthesis and antibiotic sensing, could be identified. In addition, we also identified uncharacterized enzymes which turned out to be capable of hydrolyzing β-lactam antibiotics. This technology could therefore represent a valuable tool to monitor the activity and function of other yet unexplored resistance associated enzymes in pathogenic bacteria and help to discover new drug targets for customized therapeutic interventions.